Plot the posterior predictive distribution for a simmr run
Source:R/posterior_predictive.simmr_output.R
posterior_predictive.RdThis function takes the output from simmr_mcmcand plots the
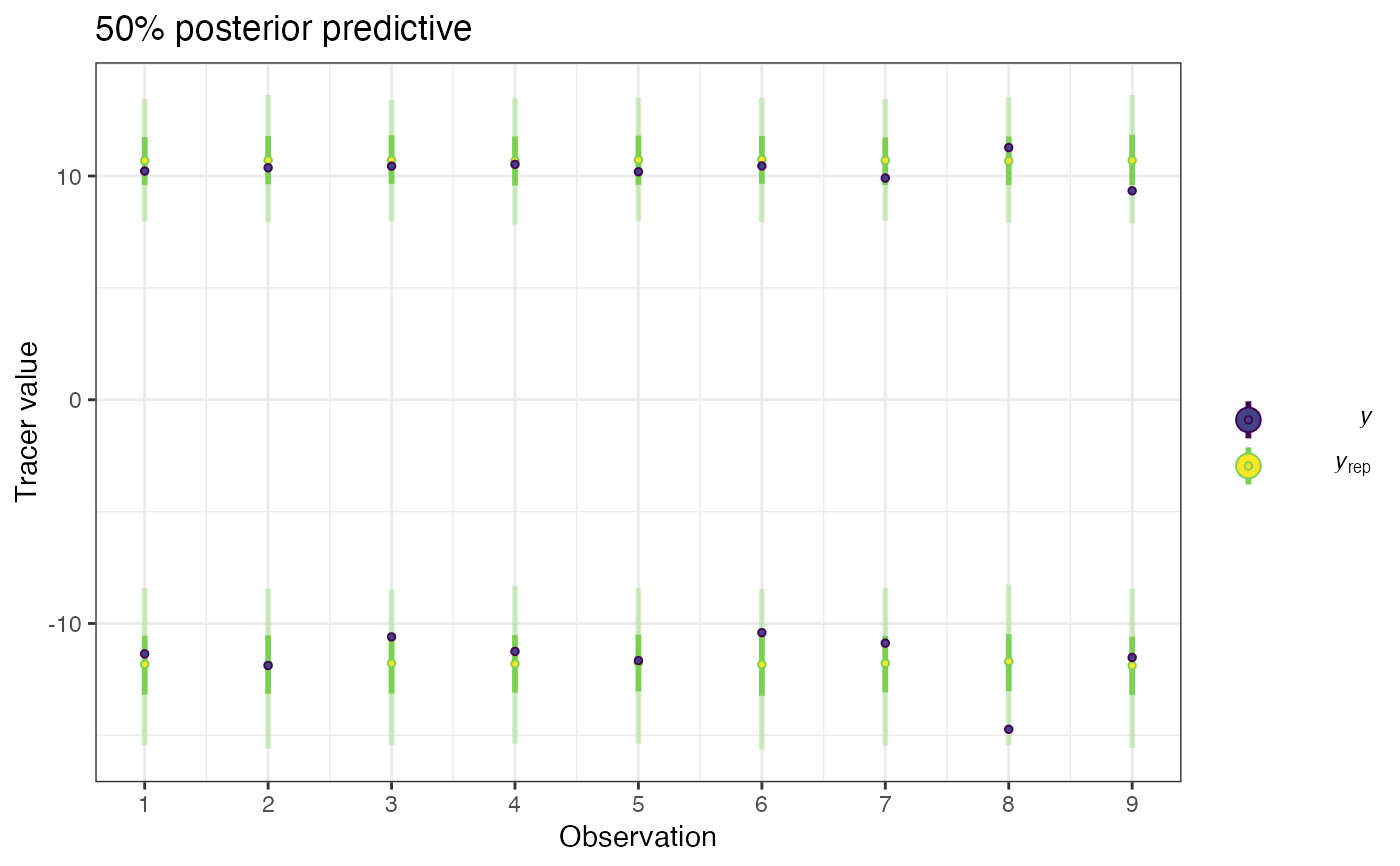
posterior predictive distribution to enable visualisation of model fit.
The simulated posterior predicted values are returned as part of the
object and can be saved for external use
posterior_predictive(simmr_out, group = 1, prob = 0.5, plot_ppc = TRUE)Arguments
- simmr_out
A run of the simmr model from
simmr_mcmc- group
Which group to run it for (currently only numeric rather than group names)
- prob
The probability interval for the posterior predictives. The default is 0.5 (i.e. 50pc intervals)
- plot_ppc
Whether to create a bayesplot of the posterior predictive or not.
Value
plot of posterior predictives and simulated values
Examples
# \donttest{
data(geese_data_day1)
simmr_1 <- with(
geese_data_day1,
simmr_load(
mixtures = mixtures,
source_names = source_names,
source_means = source_means,
source_sds = source_sds,
correction_means = correction_means,
correction_sds = correction_sds,
concentration_means = concentration_means
)
)
# Plot
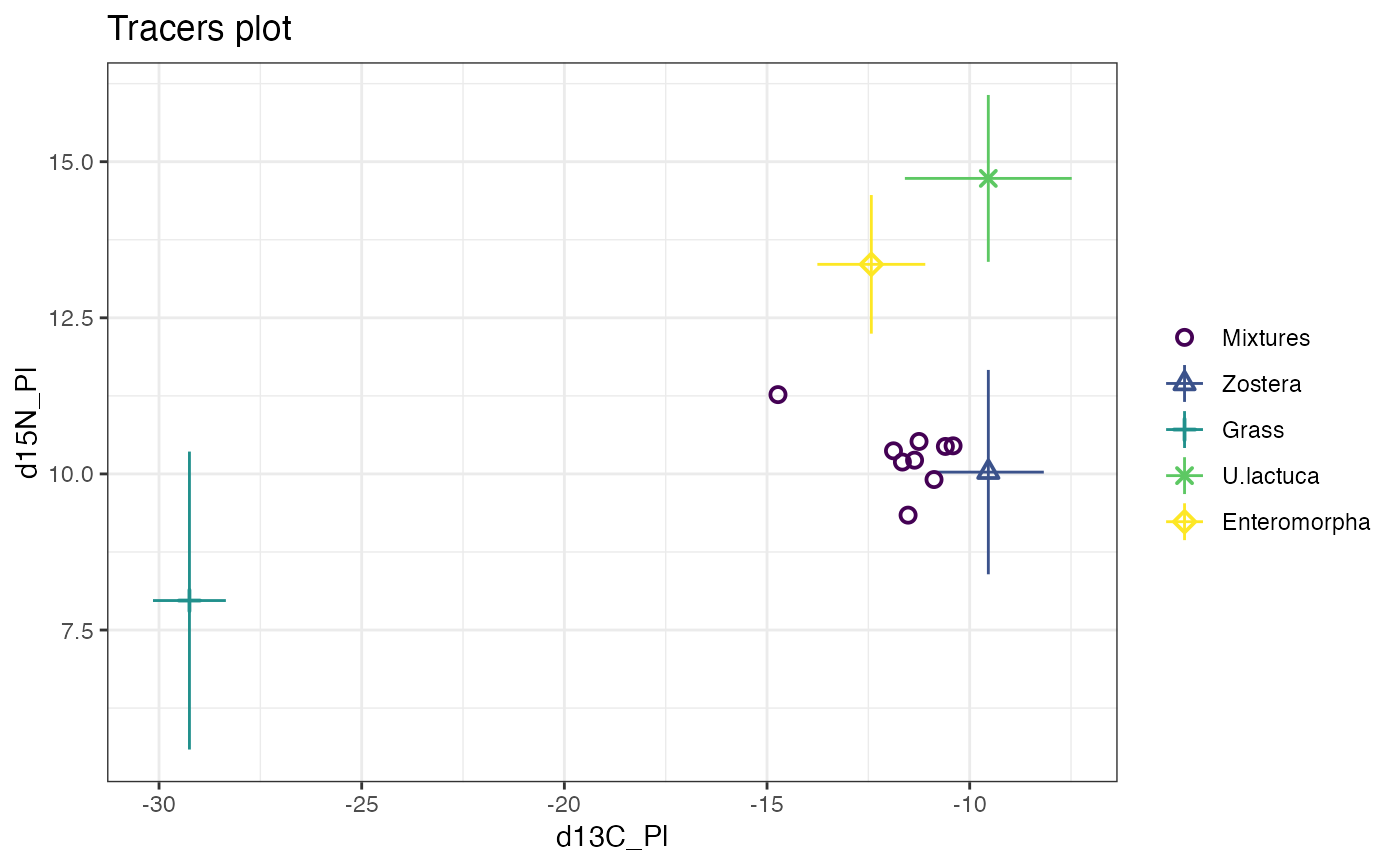
plot(simmr_1)
 # Print
simmr_1
#> This is a valid simmr input object with
#> 9 observations,
#> 2 tracers, and
#> 4 sources.
#> The source names are:
#> [1] "Zostera" "Grass" "U.lactuca" "Enteromorpha"
#> .
#> The tracer names are:
#> [1] "d13C_Pl" "d15N_Pl"
#>
#>
# MCMC run
simmr_1_out <- simmr_mcmc(simmr_1)
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 18
#> Unobserved stochastic nodes: 6
#> Total graph size: 136
#>
#> Initializing model
#>
# Prior predictive
post_pred <- posterior_predictive(simmr_1_out)
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 18
#> Unobserved stochastic nodes: 24
#> Total graph size: 154
#>
#> Initializing model
#>
# Print
simmr_1
#> This is a valid simmr input object with
#> 9 observations,
#> 2 tracers, and
#> 4 sources.
#> The source names are:
#> [1] "Zostera" "Grass" "U.lactuca" "Enteromorpha"
#> .
#> The tracer names are:
#> [1] "d13C_Pl" "d15N_Pl"
#>
#>
# MCMC run
simmr_1_out <- simmr_mcmc(simmr_1)
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 18
#> Unobserved stochastic nodes: 6
#> Total graph size: 136
#>
#> Initializing model
#>
# Prior predictive
post_pred <- posterior_predictive(simmr_1_out)
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 18
#> Unobserved stochastic nodes: 24
#> Total graph size: 154
#>
#> Initializing model
#>
 # }
# }