Summarises the output created with simmr_mcmc or
simmr_ffvb
Source: R/summary.simmr_output.R
summary.simmr_output.RdProduces textual summaries and convergence diagnostics for an object created
with simmr_mcmc or simmr_ffvb. The different
options are: 'diagnostics' which produces Brooks-Gelman-Rubin diagnostics
to assess MCMC convergence, 'quantiles' which produces credible intervals
for the parameters, 'statistics' which produces means and standard
deviations, and 'correlations' which produces correlations between the
parameters.
Arguments
- object
An object of class
simmr_outputproduced by the functionsimmr_mcmcorsimmr_ffvb- type
The type of output required. At least none of 'diagnostics', 'quantiles', 'statistics', or 'correlations'.
- group
Which group or groups the output is required for.
- ...
Not used
Value
A list containing the following components:
- gelman
The convergence diagnostics
- quantiles
The quantiles of each parameter from the posterior distribution
- statistics
The means and standard deviations of each parameter
- correlations
The posterior correlations between the parameters
Note that this object is reported silently so will be discarded unless the function is called with an object as in the example below.
Details
The quantile output allows easy calculation of 95 per cent credible
intervals of the posterior dietary proportions. The correlations, along with
the matrix plot in plot.simmr_output allow the user to judge
which sources are non-identifiable. The Gelman diagnostic values should be
close to 1 to ensure satisfactory convergence.
When multiple groups are included, the output automatically includes the results for all groups.
See also
See simmr_mcmc and simmr_ffvbfor
creating objects suitable for this function, and many more examples.
See also simmr_load for creating simmr objects,
plot.simmr_input for creating isospace plots,
plot.simmr_output for plotting output.
Examples
# \donttest{
# A simple example with 10 observations, 2 tracers and 4 sources
# The data
data(geese_data_day1)
simmr_1 <- with(
geese_data_day1,
simmr_load(
mixtures = mixtures,
source_names = source_names,
source_means = source_means,
source_sds = source_sds,
correction_means = correction_means,
correction_sds = correction_sds,
concentration_means = concentration_means
)
)
# Plot
plot(simmr_1)
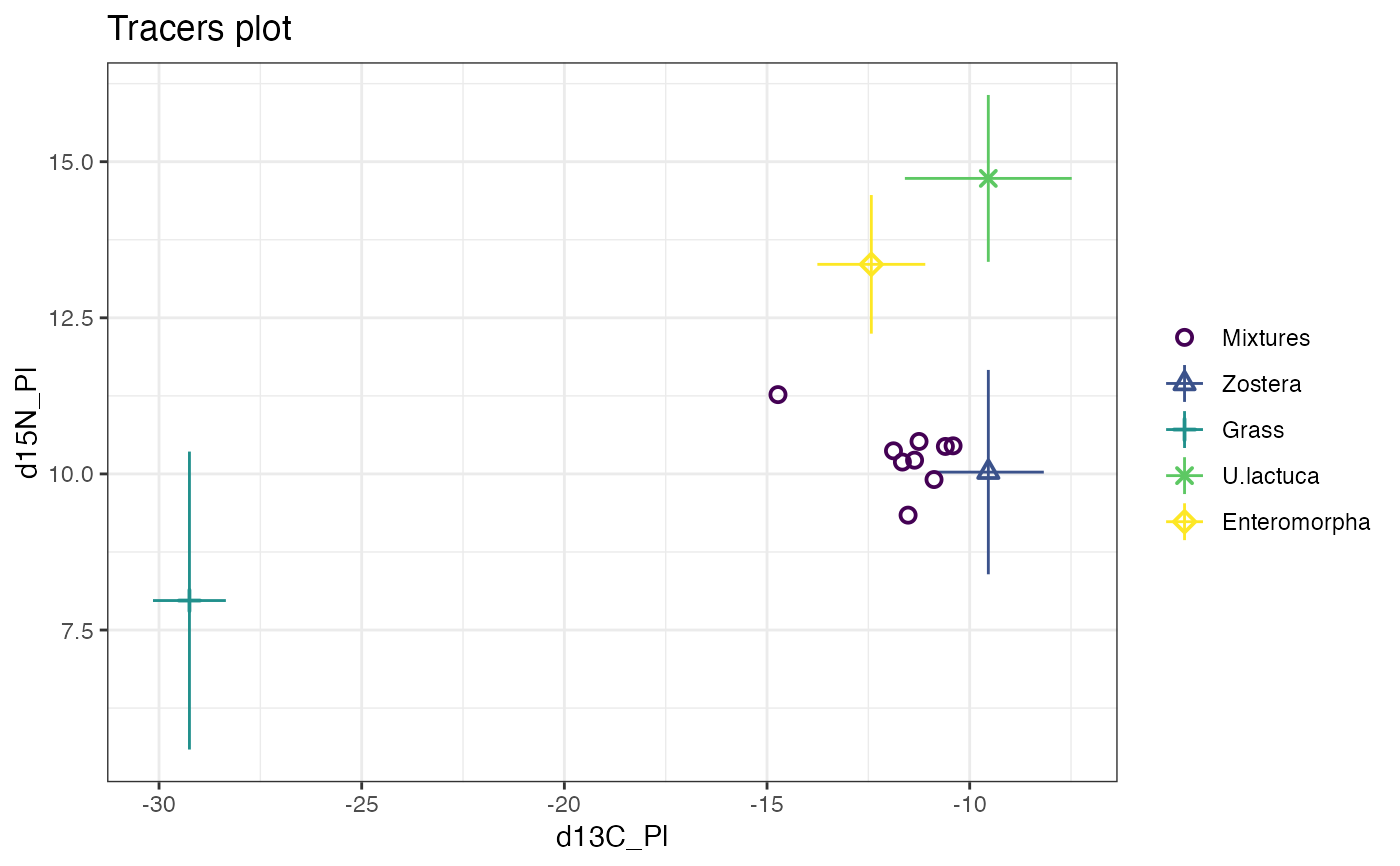 # MCMC run
simmr_1_out <- simmr_mcmc(simmr_1)
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 18
#> Unobserved stochastic nodes: 6
#> Total graph size: 136
#>
#> Initializing model
#>
# Summarise
summary(simmr_1_out) # This outputs all the summaries
#>
#> Summary for 1
#> R-hat values - these values should all be close to 1.
#> If not, try a longer run of simmr_mcmc.
#> deviance Zostera Grass U.lactuca Enteromorpha sd[d13C_Pl]
#> 1 1 1 1 1 1
#> sd[d15N_Pl]
#> 1
#> 2.5% 25% 50% 75% 97.5%
#> deviance 52.910 56.447 59.506 63.327 74.737
#> Zostera 0.333 0.505 0.583 0.665 0.807
#> Grass 0.028 0.058 0.074 0.092 0.137
#> U.lactuca 0.020 0.078 0.133 0.208 0.363
#> Enteromorpha 0.023 0.093 0.166 0.266 0.488
#> sd[d13C_Pl] 0.549 1.109 1.527 2.104 3.969
#> sd[d15N_Pl] 0.246 0.660 0.952 1.347 2.691
#> mean sd
#> deviance 60.552 5.645
#> Zostera 0.582 0.121
#> Grass 0.076 0.028
#> U.lactuca 0.150 0.094
#> Enteromorpha 0.192 0.126
#> sd[d13C_Pl] 1.717 0.909
#> sd[d15N_Pl] 1.085 0.642
#> deviance Zostera Grass U.lactuca Enteromorpha sd[d13C_Pl]
#> deviance 1.000 -0.273 0.133 0.164 0.111 0.673
#> Zostera -0.273 1.000 0.037 -0.408 -0.666 -0.106
#> Grass 0.133 0.037 1.000 0.116 -0.342 0.246
#> U.lactuca 0.164 -0.408 0.116 1.000 -0.379 0.002
#> Enteromorpha 0.111 -0.666 -0.342 -0.379 1.000 0.046
#> sd[d13C_Pl] 0.673 -0.106 0.246 0.002 0.046 1.000
#> sd[d15N_Pl] 0.684 -0.196 -0.103 0.150 0.099 0.029
#> sd[d15N_Pl]
#> deviance 0.684
#> Zostera -0.196
#> Grass -0.103
#> U.lactuca 0.150
#> Enteromorpha 0.099
#> sd[d13C_Pl] 0.029
#> sd[d15N_Pl] 1.000
summary(simmr_1_out, type = "diagnostics") # Just the diagnostics
#>
#> Summary for 1
#> R-hat values - these values should all be close to 1.
#> If not, try a longer run of simmr_mcmc.
#> deviance Zostera Grass U.lactuca Enteromorpha sd[d13C_Pl]
#> 1 1 1 1 1 1
#> sd[d15N_Pl]
#> 1
# Store the output in an
ans <- summary(simmr_1_out,
type = c("quantiles", "statistics")
)
#>
#> Summary for 1
#> 2.5% 25% 50% 75% 97.5%
#> deviance 52.910 56.447 59.506 63.327 74.737
#> Zostera 0.333 0.505 0.583 0.665 0.807
#> Grass 0.028 0.058 0.074 0.092 0.137
#> U.lactuca 0.020 0.078 0.133 0.208 0.363
#> Enteromorpha 0.023 0.093 0.166 0.266 0.488
#> sd[d13C_Pl] 0.549 1.109 1.527 2.104 3.969
#> sd[d15N_Pl] 0.246 0.660 0.952 1.347 2.691
#> mean sd
#> deviance 60.552 5.645
#> Zostera 0.582 0.121
#> Grass 0.076 0.028
#> U.lactuca 0.150 0.094
#> Enteromorpha 0.192 0.126
#> sd[d13C_Pl] 1.717 0.909
#> sd[d15N_Pl] 1.085 0.642
# }
# MCMC run
simmr_1_out <- simmr_mcmc(simmr_1)
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 18
#> Unobserved stochastic nodes: 6
#> Total graph size: 136
#>
#> Initializing model
#>
# Summarise
summary(simmr_1_out) # This outputs all the summaries
#>
#> Summary for 1
#> R-hat values - these values should all be close to 1.
#> If not, try a longer run of simmr_mcmc.
#> deviance Zostera Grass U.lactuca Enteromorpha sd[d13C_Pl]
#> 1 1 1 1 1 1
#> sd[d15N_Pl]
#> 1
#> 2.5% 25% 50% 75% 97.5%
#> deviance 52.910 56.447 59.506 63.327 74.737
#> Zostera 0.333 0.505 0.583 0.665 0.807
#> Grass 0.028 0.058 0.074 0.092 0.137
#> U.lactuca 0.020 0.078 0.133 0.208 0.363
#> Enteromorpha 0.023 0.093 0.166 0.266 0.488
#> sd[d13C_Pl] 0.549 1.109 1.527 2.104 3.969
#> sd[d15N_Pl] 0.246 0.660 0.952 1.347 2.691
#> mean sd
#> deviance 60.552 5.645
#> Zostera 0.582 0.121
#> Grass 0.076 0.028
#> U.lactuca 0.150 0.094
#> Enteromorpha 0.192 0.126
#> sd[d13C_Pl] 1.717 0.909
#> sd[d15N_Pl] 1.085 0.642
#> deviance Zostera Grass U.lactuca Enteromorpha sd[d13C_Pl]
#> deviance 1.000 -0.273 0.133 0.164 0.111 0.673
#> Zostera -0.273 1.000 0.037 -0.408 -0.666 -0.106
#> Grass 0.133 0.037 1.000 0.116 -0.342 0.246
#> U.lactuca 0.164 -0.408 0.116 1.000 -0.379 0.002
#> Enteromorpha 0.111 -0.666 -0.342 -0.379 1.000 0.046
#> sd[d13C_Pl] 0.673 -0.106 0.246 0.002 0.046 1.000
#> sd[d15N_Pl] 0.684 -0.196 -0.103 0.150 0.099 0.029
#> sd[d15N_Pl]
#> deviance 0.684
#> Zostera -0.196
#> Grass -0.103
#> U.lactuca 0.150
#> Enteromorpha 0.099
#> sd[d13C_Pl] 0.029
#> sd[d15N_Pl] 1.000
summary(simmr_1_out, type = "diagnostics") # Just the diagnostics
#>
#> Summary for 1
#> R-hat values - these values should all be close to 1.
#> If not, try a longer run of simmr_mcmc.
#> deviance Zostera Grass U.lactuca Enteromorpha sd[d13C_Pl]
#> 1 1 1 1 1 1
#> sd[d15N_Pl]
#> 1
# Store the output in an
ans <- summary(simmr_1_out,
type = c("quantiles", "statistics")
)
#>
#> Summary for 1
#> 2.5% 25% 50% 75% 97.5%
#> deviance 52.910 56.447 59.506 63.327 74.737
#> Zostera 0.333 0.505 0.583 0.665 0.807
#> Grass 0.028 0.058 0.074 0.092 0.137
#> U.lactuca 0.020 0.078 0.133 0.208 0.363
#> Enteromorpha 0.023 0.093 0.166 0.266 0.488
#> sd[d13C_Pl] 0.549 1.109 1.527 2.104 3.969
#> sd[d15N_Pl] 0.246 0.660 0.952 1.347 2.691
#> mean sd
#> deviance 60.552 5.645
#> Zostera 0.582 0.121
#> Grass 0.076 0.028
#> U.lactuca 0.150 0.094
#> Enteromorpha 0.192 0.126
#> sd[d13C_Pl] 1.717 0.909
#> sd[d15N_Pl] 1.085 0.642
# }